Disclaimer: Early release articles are not considered as final versions. Any changes will be reflected in the online version in the month the article is officially released.
Author affiliations: Institute for Food Safety and Hygiene, Section of Veterinary Bacteriology, Vetsuisse Faculty University of Zurich, Zurich, Switzerland (G. Ghielmetti, U. Friedel, R. Stephan); Bavarian Health and Food Safety Authority, Oberschleissheim, Germany (A.M. Kupca, J.M. Riehm); Institute for Veterinary Disease Control, Austrian Agency for Health and Food Safety (AGES), Innsbruck and Mödling, Austria (H. Weinberger, S. Revilla-Fernández, E. Hofer, W. Glawischnig)
Tuberculosis (TB) is one of the most prevalent zoonotic diseases worldwide and remains the leading cause of death from a single infectious agent (1). The causative pathogens of TB in humans and animals are a group of closely related acid-fast bacilli commonly known as the Mycobacterium tuberculosis complex (MTBC). One animal-adapted sublineage within the complex, M. microti, was first isolated from field voles (Microtus agrestis) that had granulomatous tuberculosis-like lesions (2). Although wild rodents, such as bank voles (Myodes glareolus), wood mice (Apodemus sylvaticus), and shrews (Sorex araneus), are considered to be primary reservoirs for M. microti, several other hosts have been identified, including domestic and wild animals (3,4). Overall, cats (5,6), New World camelids (7), and free-ranging wild boar (8–10) seem to be prone to M. microti infections; humans (11–14) and other animal species, including pigs (15), goats (16), cattle (17,18), dogs (19), captive meerkats (20), squirrel monkeys (21), and ferrets (14), are most likely incidental hosts.
This broad host range, however, highlights the pathogenic potential of M. microti and the need to reveal its virulence mechanisms. Comparative genomics studies have identified >100 genes whose presence are facultative and differ among members of MTBC. Many of these genes occur in chromosomal regions of difference (RD) that have been deleted from certain species and that may confer differences in phenotype, host range, and virulence (22). Isolates of the animal-adapted ecotype defined as M. microti are characterized by the deletion of the RD1mic in the RD1 region, which includes open reading frame coding for well-known virulence factors, such as early secreted antigenic target (ESAT) 6, locus tag Rv3875, and CFP-10, a culture filtrate protein encoded by the neighboring gene Rv3874 (23). Strains lacking RD1 are likely to be less virulent or pathogenic than other members of the MTBC possessing an intact locus (22). However, pulmonary and disseminated M. microti infections have been described in both immunocompromised and immunocompetent human patients in different countries in Europe (11,12,14,24). Until recently, reports of M. microti infections were geographically restricted to continental Europe and the United Kingdom. However, a recent study from South Africa revealed the presence of this Mycobacterium species in 1.9% of local human tuberculosis cases (25). These findings highlight the potential of M. microti to cause clinical illness in immunocompetent patients and suggest that the pathogenicity of certain strains is higher than previously estimated. Therefore, it is crucial to identify clinical MTBC isolates at the species level, and the zoonotic risk posed by M. microti should be further evaluated.
The mode of infection of M. microti can only be speculated for humans, animals, and in particular herbivores, such as free-ranging red deer. Similar to that for M. caprae, transmission of M. microti is likely to occur indirectly through a contaminated environment. Wounds in the oral cavity may play an important role as entry ports for M. microti; involvement of the lungs, heart, and eventually additional organs is most likely a consequence of bacteremia, as it was in other animal species (3,5). The first confirmed M. caprae TB case in deer in western Austria was recorded in 1998. Subsequent infections in cattle, deer, and humans were reported in the same area (26). As a consequence, an ongoing wildlife surveillance program monitoring M. caprae in deer was started in 2008 (27). Furthermore, Germany in 2007 and Switzerland in 2013 reported anecdotal outbreaks in cattle (28,29). During 2011–2013, a monitoring program coordinated by the EMIDA ERA-Net (Coordination of European Research on Emerging and Major Infectious Diseases of Livestock European Research Area Networks) partnership and including specific regions of Austria, Switzerland, Germany, and Italy was conducted with the aim of investigating the prevalence of bovine tuberculosis (bTB) in red deer and additional wildlife species such as wild boar, chamois, and roe deer (30,31). We report 3 TB cases in red deer identified within the framework of these monitoring programs.
Study Material and Methods
Cases
Three cases of natural M. microti infections in red deer were identified (Table 1). The deer in case 1, a highly emaciated 9-year-old stag from the province of Vorarlberg, Austria, was humanely killed by a local game warden, who submitted the lungs, heart, and lymphatic tissues (including medial retropharyngeal, tracheobronchial, and mediastinal lymph nodes) fresh for pathoanatomic inspection. Thereafter, histologic examination and mycobacterial analysis of the lungs were performed. The deer in case 2 was a stag 1–3 years of age and in case 3 a hind >2 years of age, both in the province of Miesbach, Germany, where deer are regularly hunted. The heads, lungs, intestines, and associated lymph nodes were macroscopically inspected; subsequently, a histopathologic and bacteriologic examination of the lungs and lymph nodes were performed.
Mycobacterial Analyses and Histologic Examination
We isolated mycobacteria following a standardized protocol as described elsewhere (32). In brief, 2–3 g of minced tissue samples were homogenized in 5 mL 0.9% NaCl solution by using a rotating-blade macerator system (Ultra-Turrax IKA, https://www.ika.com). The suspension was decontaminated by using 1% N-acetyl-L-cystein-NaOH solution and neutralized with 20 mL phosphate buffer (pH 6.8). We centrifuged the solution for 20 min at 3300 × g and plated the obtained pellet on 2 growth media: Löwenstein-Jensen medium with glycerin and PACT (polymyxin B, amphotericin B, carbenicillin, and trimethoprim) and Stonebrink medium with pyruvate and PACT (BD, https://www.bd.com). Cultures of lung and lymph node specimens on solid Stonebrink medium yielded growth of suspicious mycobacterial colonies after 4–6 wk of incubation at 37°C. The isolates were identified by using GenoType MTBC reversed line blotting (Hain Lifescience, https://www.hain-lifescience.de). For histologic examination, we fixed tissue samples in 10% nonbuffered formalin for ≈48 h, then trimmed and routinely embedded them in paraffin wax. Sections of 3–4 μm were prepared and stained with hematoxylin and eosin (HE) and Ziehl Neelsen (ZN) or modified ZN (33).
Investigation of Phylogenetic Relationships
DVR spoligotyping (direct variable repeat spacer oligonucleotide typing) was performed using a commercial microarray system (Alere Technologies, https://www.globalpointofcare.abbott) with integrated data analysis as described elsewhere (29). Multilocus variable-number tandem repeats analysis (MLVA) was conducted based on the 24-loci panel standardized for M. tuberculosis typing (34). We amplified the single markers by endpoint PCR and subsequently analyzed them by using a capillary electrophoresis device (29). To investigate the phylogenetic relationships between the 3 isolates from red deer, we analyzed 8 additional strains isolated from different wild and domestic hosts that originated from the regions bordering Germany, Austria, and Switzerland by MLVA (Table 2). We calculated a neighbor-joining phylogenic tree based on the copy numbers of the tested loci using the MIRU-VNTRplus (https://www.miru-vntrplus.org/MIRU) server and exported it using MEGAX version 10.11 (35).
Results
Case 1
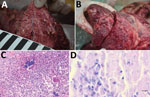
Postmortem examination of the stag revealed multiple enlarged lymph nodes exhibiting a whitish cut surface. The lung tissue showed severe pyogranulomatous pleuropneumonia with multifocal to confluent cavernous granulomas of 2–10 mm diameter (Figure 1, panel A). Multifocal to coalescing granulomas of 4–25 mm diameter were observed on the surface of the epicardium (Figure 1, panel B). Histopathologic examination of the lung revealed a severe chronic multifocal to coalescing pyogranulomatous pneumonia with focal areas of fibrosis, central areas of necrosis and mineralization, surrounded by numerous epithelioid macrophages and a few multinucleated Langhans giant cells. Lymphocytes, plasma cells and occasionally well-differentiated fibroblasts surrounded the granulomas (Figure 1, panel C). Few extracellular and intracellular acid-fast bacilli were identified in the pulmonary lesions by using ZN staining (Figure 1, panel D).
Case 2
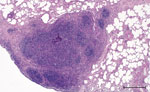
Macroscopically, a single yellowish, pinhead-sized focus in the left dorsal main lobe of the lung of this stag was observed. Lymph nodes and intestines did not display any abnormalities. Histologically, the pulmonary focus consisted of macrophages and lymphocytes with single multinucleated Langhans-type giant cells in the lesion, surrounded by eosinophilic lymphocytes (Figure 2). Numerous eosinophilic granulocytes were seen in the pulmonary lymph node. These findings were compatible with a parasitic infestation. Intracellular acid-fast bacilli could not be identify by using modified ZN staining.
Case 3
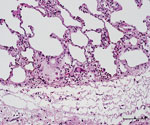
The caudal part of the main lobes of the lung of this hind showed multiple whitish foci <0.5 cm in size. Enlarged pulmonary lymph nodes and multifocal fibroblastic pleuritis were observed. The histologic examination revealed moderate purulent bronchitis and bronchiolitis with several intraluminal stages of lungworms and infiltration of numerous eosinophilic granulocytes. We observed very few multinucleated Langhans-type giant cells (Figure 3) and could identify no intracellular acid-fast bacilli in the lesions using modified ZN staining.
Investigation of Phylogenetic Relationships
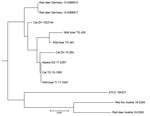
The isolates from 11 animals (3 wild boars, 3 cats, 1 alpaca, and 1 red fox), integrated for further comparative genotyping, exhibited the same spoligotype signature, SB0118, characterized by the presence of spacers 37–38 (https://www.mbovis.org). The same signature is also registered in the international spoligotyping database SpolDB4 as ST 539 and is characteristic of M. microti (36). MLVA showed 2 distinct genotypes (Figure 4; Appendix), 1 for the 2 identical isolates from the red deer from Germany and 1 for the red deer isolate from Austria. Of interest, the isolates from Germany were closely related to isolates from Switzerland, whereas 2 isolates from Austria, originating from a red deer (case 1) and a red fox, were genetically more distant despite their geographic proximity (Figure 5).
Discussion
Case 1 in this study reported a tuberculosis-positive stag killed in the alpine region in western Austria manifesting clinical signs of tuberculosis. Tuberculosis caused by M. caprae has been described several times in domestic animals and wildlife in this area (26,30). Specifically, red deer represent a reservoir and a possible source of infection with M. caprae for cattle in Austria, Germany, Italy, and Switzerland (26,28,29,37). M. microti has been isolated only once in Austria, from a red fox without visible lesions (Table 2). This fox was located at a distance of ≈30 km from the site in Germany where the M. microti–positive stag in case 1 was found. Clear evidence proving transmission of M. microti between individual animals of the same species or between species is missing. Human-to-human transmission regarding this pathogen has previously been investigated and the possibility cannot be dismissed (14). However, ingesting feed or water from contaminated sources, for example, might play an important role in transmitting mycobacteria to wildlife. In fact, recent reports suggest that M. microti infections might often occur through oral ingestion and that direct transmission between animals is less likely (9,20).
The presence of MTBC in wild red deer seems to depend on multiple factors, such as population density, TB prevalence in nearby cattle or other wildlife species, and the morphologic structure of the habitat. Observations made from infected wild deer in New Zealand showed that M. bovis prevalence decreased substantially after control of TB-infected possums, suggesting that wild deer may be spillover hosts that can be regularly reinfected by possums (38). In fact, considering the high levels (>50%) of bTB in cattle in Europe before eradication campaigns, sporadic transmissions to wildlife populations might have occurred. It is however surprising to note that by successfully lowering the prevalence in cattle, the disease has been eradicated from wild deer populations, such as in Switzerland (38). The situation is considerably different for deer in captivity or in high-density populations, which could be the origin of TB dissemination to other species (39). Of interest, the virulence of M. microti seems to vary greatly both between host species and within the same species. Most visible lesions compatible with TB diagnosed in M. bovis– and M. caprae–infected red deer are located in the lymph nodes, particularly the medial retropharyngeal and mesenteric lymph nodes, suggesting oral rather than aerosol transmission. The respiratory tract, including the lungs and associated lymph nodes, seems to be affected by MTBC in a secondary phase of the infection, which is also likely for M. microti infections in red deer (30).
DVR spoligotyping analysis is a popular technique worldwide for molecular characterization of MTBC of animal origin with excellent resolution and cost-benefit ratio (40,41), but the discriminatory power is too low to prove any link on an epidemiologic level or even minute transmission patterns among M. microti lineages. The molecular background of M. microti seems highly conserved, and traceback analyses are delicate. The 11 isolates included in this study were collected over an 8-year period from regions bordering Austria, Germany, and Switzerland (Table 2). These strains originated from 5 different wild and domestic animal species, and most of the animals in these cases showed severe TB lesions. Although the number of isolates investigated was small, in accordance with previous studies, no correlation between host species and M. microti genotypes was observed (4,42). Moreover, even though the isolates from Switzerland were genetically close, genetic variation determined by MLVA did not correlate with the relative geographic distance of their origin. In fact, the isolates from red deer in Germany were genetically closer to the strains from Switzerland, whereas the 2 isolates from Austria, originating in the border region shared with Switzerland, were genetically more distant (Figures 4, 5), which suggests that the circulation of host-adapted M. microti genetic lineages is unlikely. MLVA has been successfully used worldwide as an ancillary tool for animal TB epidemiologic surveillance and outbreak investigations in multihost scenarios (27,43–45). However, the discriminatory capacity of whole-genome sequencing has elsewhere been shown to be superior for identifying MTBC strains belonging to the same regional clonal complex, which may apply to M. microti as well (39,46). Under certain specific circumstances, such as for formalin-fixed, paraffin-embedded samples or extremely fastidious strains, MLVA represents a valid alternative to whole-genome sequencing.
These findings highlight the wide host range of M. microti and suggest that it might be an opportunistic pathogen rather than a host-adapted MTBC member, such as M. tuberculosis. In the past, similar to Mycobacterium bovis BCG strains, certain vole strains of M. microti have been used to develop live attenuated human TB vaccines in the United Kingdom and the former Czechoslovakia (47–49). Therefore, marked virulence differences between M. microti strains are likely to exist (50).
Some seemingly feasible theories about the natural transmission route of M. microti suggest that the natural foci and reservoirs of this animal-adapted lineage are small rodents and that the pathogen subsequently infects predators, such as cats or foxes, through ingestion; the mode of infection for herbivores, such as red deer or alpacas, remains ill defined. The lesions we observed in the lungs of the deer in case 1, however, provide strong evidence of bacterial shedding, which might occur either as a consequence of inflammatory processes that break into the airways or by infection of alveolar macrophages that are part of the exudate (5), which result in environmental contamination and further transmission of the pathogen (3). It is therefore alarming that animal species, such as red deer, that can cover long distances in short periods of time might contribute to the spread of M. microti, an MTBC agent.
Because of the potential zoonotic risk related to MTBC members, rapidly and accurately identifying the mycobacterial species causing disease in animals hunted for human consumption is crucial. Once MTBC is detected, determining whether M. bovis or M. caprae is present is of primary importance for veterinary and public health authorities. To date, molecular testing of cultured bacteria remains the preferred method for differentiating MTBC species. Because of the fastidious nature of M. microti and the extremely slow growth rate of specific animal strains, this differentiation can take several months or remain incomplete in cases where the mycobacterium cannot be cultured. On the basis of published data, it can be assumed that a large proportion of M. microti infections remain culture negative, even if the incubation time is prolonged to 18 weeks (8). Therefore, identifying species using molecular methods on native samples is recommended.
These findings show the morphologic versatility of lesions induced by M. microti in red deer. Given the absence of typical pulmonary lesions in some cases, such as in the red deer in cases 2 and 3, diagnostic pathologists must remain highly alert. Incidence of this pathogen should be monitored within the framework of bTB surveillance programs and suspicious cases differentiated from classical bTB caused by M. bovis and M. caprae. The actual occurrence of M. microti in wildlife may be underestimated, and personnel involved in bTB monitoring programs should be aware of its pathogenicity and zoonotic potential. Therefore, molecular methods to differentiate this member of the MTBC should be included in the diagnostic workflow of bTB reference laboratories.
Dr. Ghielmetti is a researcher at the Institute for Food Safety and Hygiene, Section of Veterinary Bacteriology, University of Zurich, Switzerland. His research interests include the molecular characterization and epidemiology of zoonotic pathogens.
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
The Link LonkJune 30, 2021 at 12:24AM
https://wwwnc.cdc.gov/eid/article/27/8/21-0634_article
Mycobacterium microti Infections in Free-Ranging Red Deer (Cervus elaphus) - CDC
https://news.google.com/search?q=Red&hl=en-US&gl=US&ceid=US:en

No comments:
Post a Comment